DIPlib 3.0.0 released
After several years of hard work, DIPlib 3 is finally out of beta.
I presented the DIPlib 3 project on this blog three and a half years ago, when I discussed the advantages of the new C++ library over its C predecessor, and especially how C++ code using DIPlib 3 is quite similar to MATLAB code using DIPimage (meaning it is simple!). I also mentioned the new Python bindings, PyDIP, which I demonstrated in last week’s blog post.
For this post I thought I’d show off the image visualization tool that my friend Wouter Caarls wrote for the DIPlib project. Prototyping Image Analysis algorithms often requires visualizing intermediate results, which makes MATLAB or Python much more attractive than C++ for prototyping, as those environments have many options for interactive visualization of data. DIPlib’s new visualization tool, DIPviewer, fills this role for C++, making prototyping Image Analysis algorithms directly in C++ quite easy and pleasant.
To visualize an image, simply call dip::viewer::Show(). Multiple images can be shown at the
same time. A call to dip::viewer::Spin() pauses execution of the program and allows interaction
with the visualized images. Here’s an example:
#include "diplib.h"
#include "diplib/simple_file_io.h"
#include "diplib/linear.h"
#include "dipviewer.h" // Add this
int main() {
dip::Image anat = dip::ImageRead( "sub-1-anat-sub-1_T1w.nii" );
dip::viewer::Show( anat, "anat image" ); // Add this
dip::Image H = dip::Hessian(anat);
dip::viewer::Show( H, "Hessian" ); // Add this
dip::viewer::Spin(); // Add this
// ... rest of program
}
The two images are displayed like this:
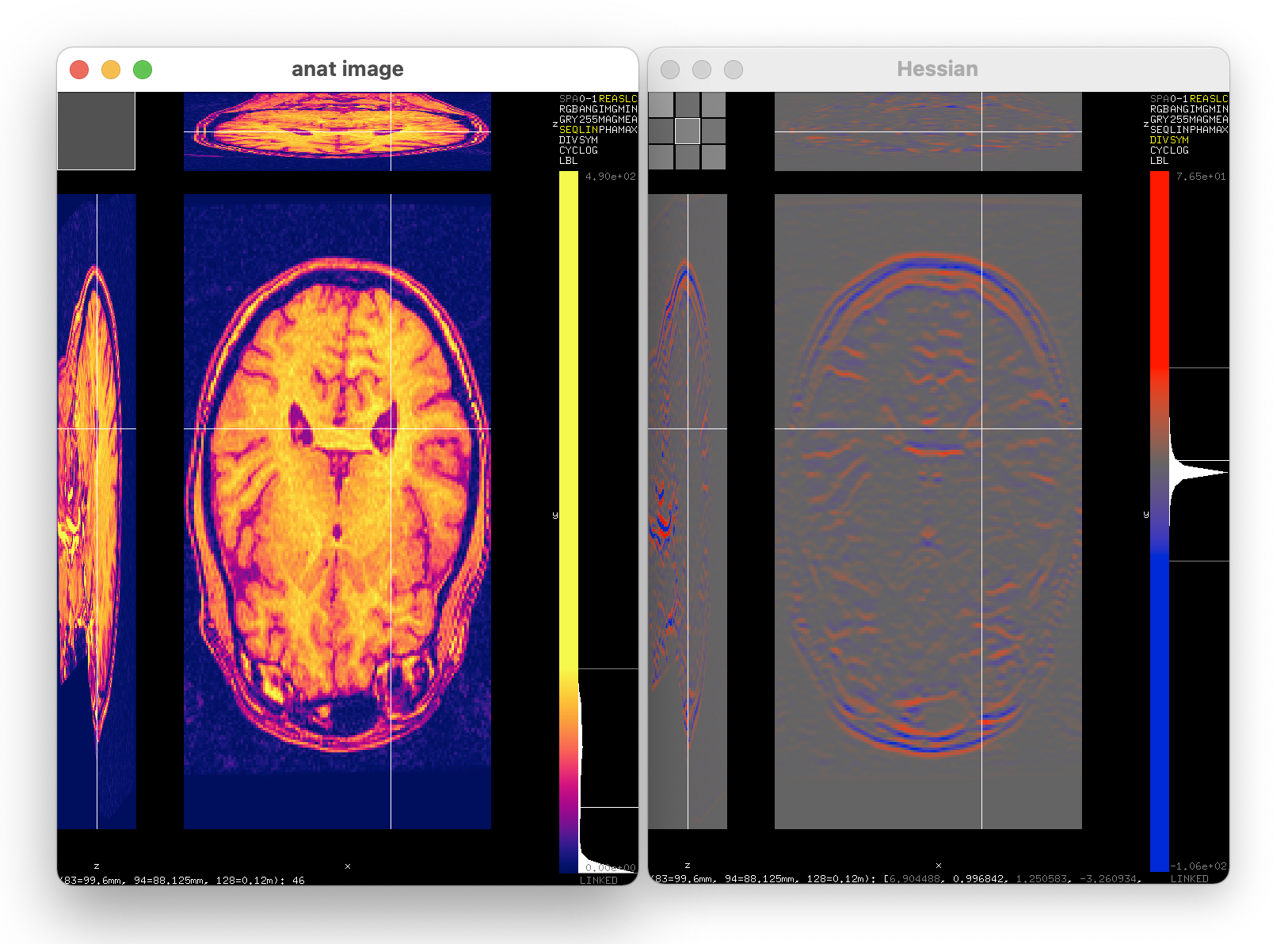
Many interesting things can be seen in the image above: the 3D images are shown as three slices through the volume, one slice is the main display, the other two slices are meant to aid navigation. The display can show images with more dimensions just as easily, and one can choose how each of these three panels slices the image. To the right is a histogram and a color bar, indicating the range of the image data and the selected range for display, which can be adjusted there. In the top-right corner are controls allowing one to adjust how the image is displayed, including the color map. At the bottom of the window one can read out the coordinates of the cursor, both in pixels and in physical units, as well as the value of the pixel under the cursor.
The image displayed in the right window, the Hessian, is a 3x3 tensor image. The top-left corner of the window contains a control allowing one to select which tensor component is visualized as gray-value image. When selecting the RGB display mode, up to three tensor components can be selected, one for each of the red, green and blue channels.
Finally, the two windows were linked, which can be seen by the cursor over the two images being at the same spot. The numbers read out at the bottom of the two windows correspond to the same pixel in the two images. When navigating in one of the windows, the other window follows along. This is incredibly convenient when examining both the input image and one or more intermediate results, where one wants to learn how the image was transformed.
Pressing Ctrl+Shift+W closes all windows and allows the program to continue execution.
One more thing: Note how the example code above loads an MRI image from a file with
an .nii extension. I had, until very recently, not heard of this file format.
Luckily, it is one of the
150 file formats
that the Bio-Formats library understands.
Turns out it is the NIfTI format used by the fMRI community.
DIPlib 3 has an interface to Bio-Formats, and so I’m able to load this file through
dip::ImageRead(). Coincidentally, the interface to Bio-Formats in DIPlib was also
written by Wouter Caarls.
Thank you Wouter!